Exome sequencing is a widely used and cost-effective targeted sequencing method.
The application is used to enrich, sequence, and analyze the coding and regulatory regions of the genome, allowing detection of disease causing candidate variants (including SNPs, insertions and deletions), population genetics, and more.
Researchers who request this service from the TGC receive the following analysis results files:
- Final report – detailed summary of the analysis, including explanations and statistics for each step, technical details, and all necessary information for publishing.
- Raw data – the sequenced reads in ‘fastq’ format.
- Genome viewer files – visualization of the aligned reads (bam files), allowing further exploration of coverage profiles and a broader view of selected variants loci.
- An organized results table – Includes detailed information of each detected variant, including genomic annotations of the variant’s locus and project-specific filtering and merging criteria, integrating data from all relevant samples.
- Coverage files – A list of low coverage regions and overall coverage profile details.
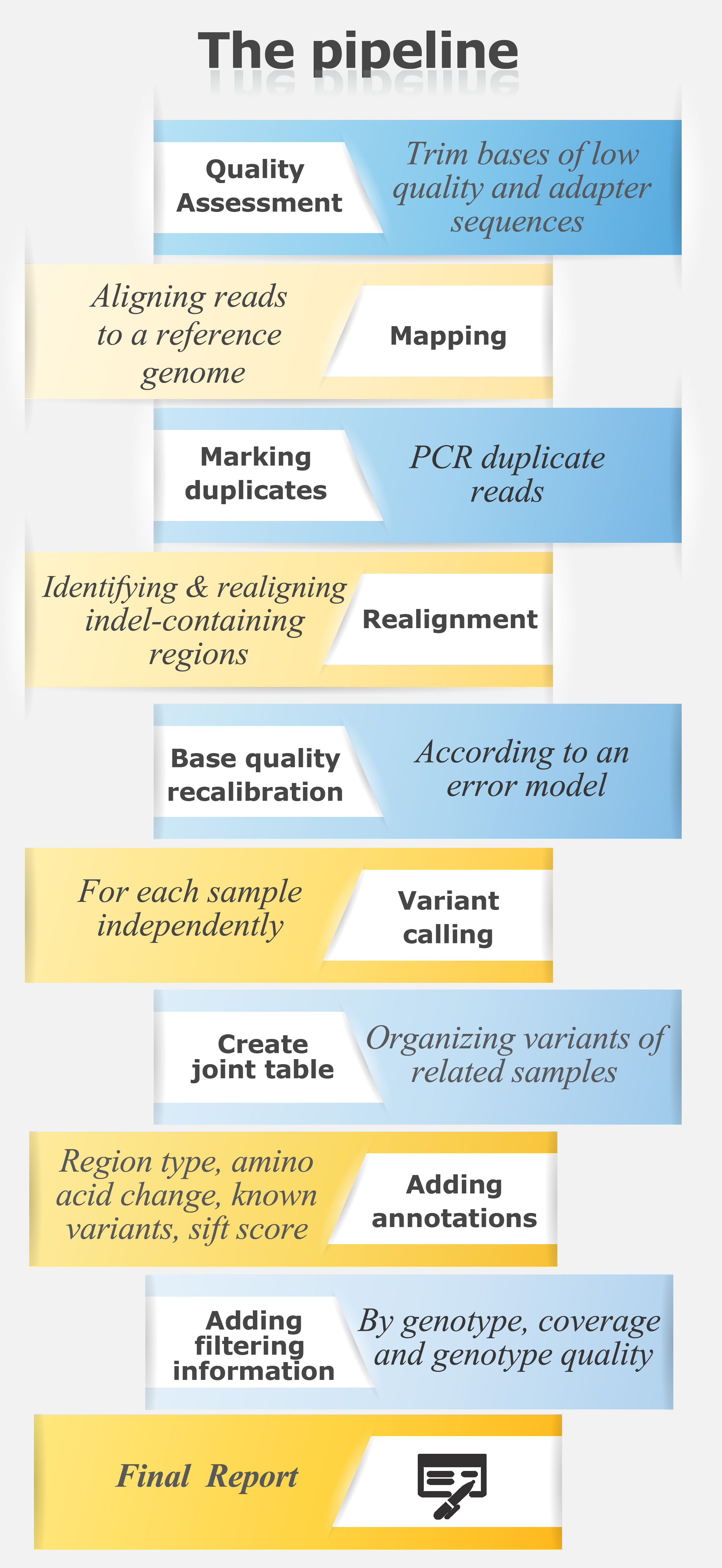
For exome sequencing pipeline in text format
Example of Exome Analysis Variants Table:
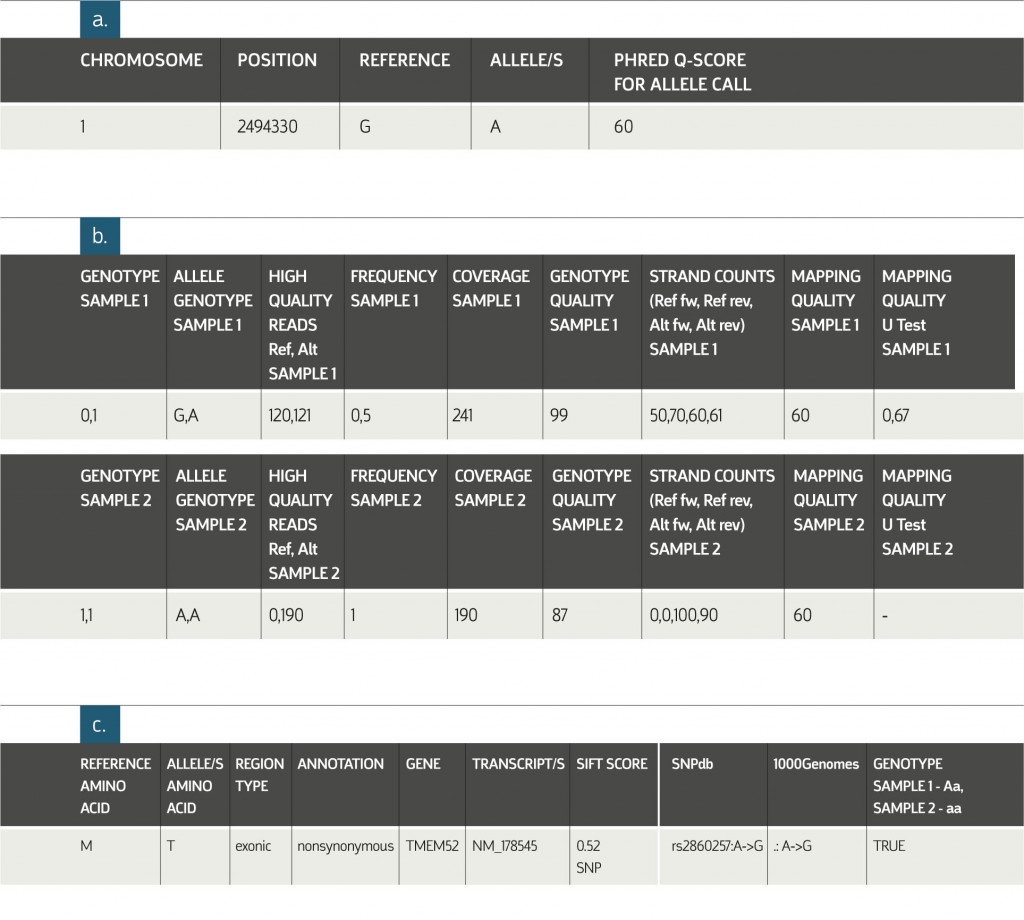 a. General information about the detected allele(s).
a. General information about the detected allele(s).
b. Sample-specific information. The columns pictured above appear in the table for each of the
analyzed samples.
c. Genomic annotations of the variant position in the genome. IDs of known variants in published databases and filtering flags. Filtering flags are project-specific; the main filtering criteria are selected based on information provided by the researcher.